|
Working with the Graph
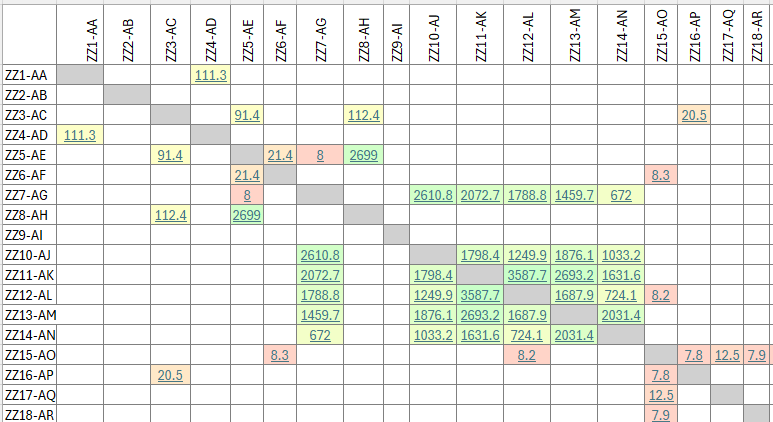
The graph will initially appear as a black graph, which may be a solidd "hairball" mass if you have a lot of kits in your project. You need to spread the graph apart so that you can see the nodes and how they connect. You also need to identify clusters and color them so that the graph allows you to visualize the clusters. You masy also need to reduce the dimensionality to include only the most-connected kits so that the graph is not overly complex. And you need to label the nodes and resize the labels and also maybe resize the edges (the connecting lines) based on how many cMs they represent.
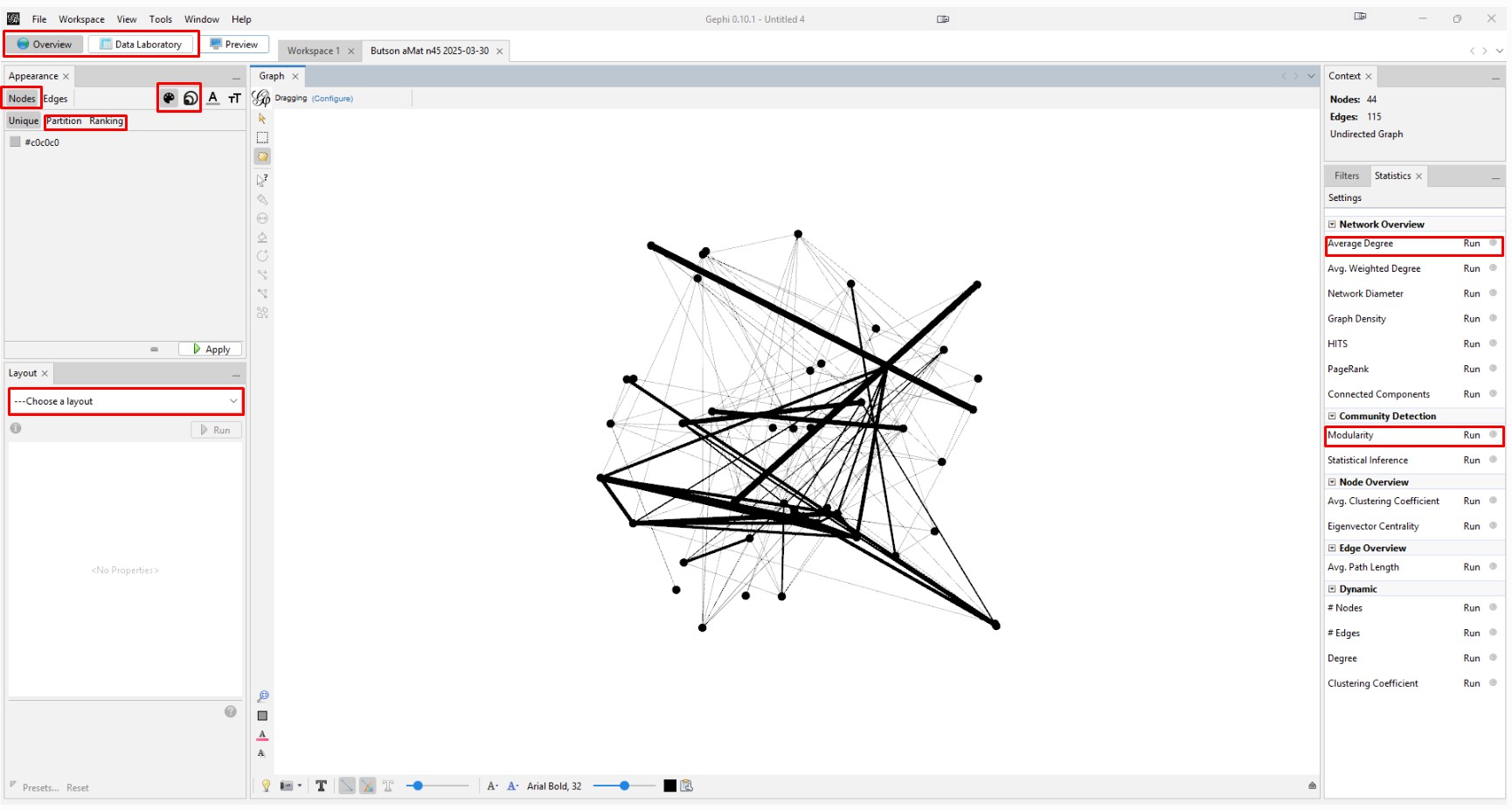
This image highlights the key places to click in the following steps. The control panel is definitely daunting because it has so many features.
Spreading the Graph Apart: The graph's overall visual shape varies depending on which "Layout" you choose in the "Overview" tab on the left side tool bar. After experimenting with different layouts, I opted for the "Fruchterman Reingold" layout with its default parameters. Choose that layout from the pulldown menu, change the "Gravity" to 2, and then click "Run" and then click "Stop". You can always recenter the graph image with the magnifying glass icon at bottom left and zoom in or our with the scroll wheel on your mouse. Zooming does focus on where on the graph you hover your cursor.
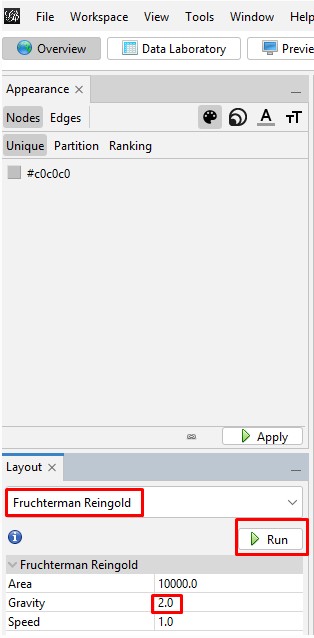
| Run then Stop
| 
|
Reducing the Dimensionality: Since this case has only 45 kits, dimensionality reduction is not necessary. But if you have a real "hairball" you will need to reduce the dimensionality so you can focus on the most connected kits. I explain the steps to do this in my instructions on my web page on generations matrix graphs.
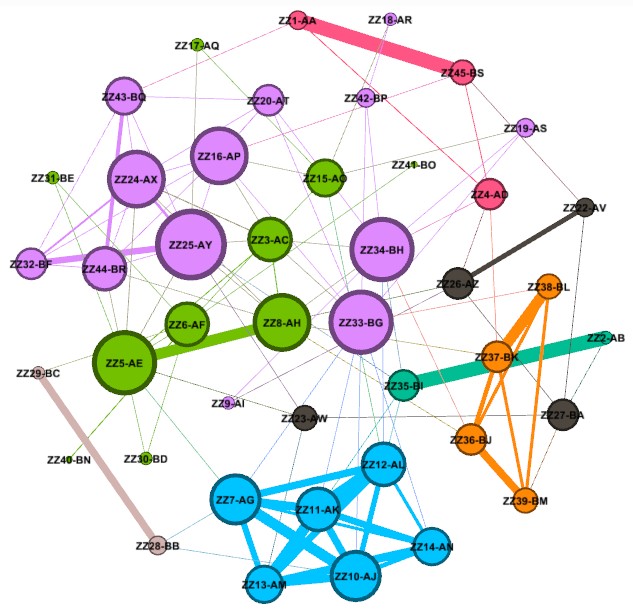
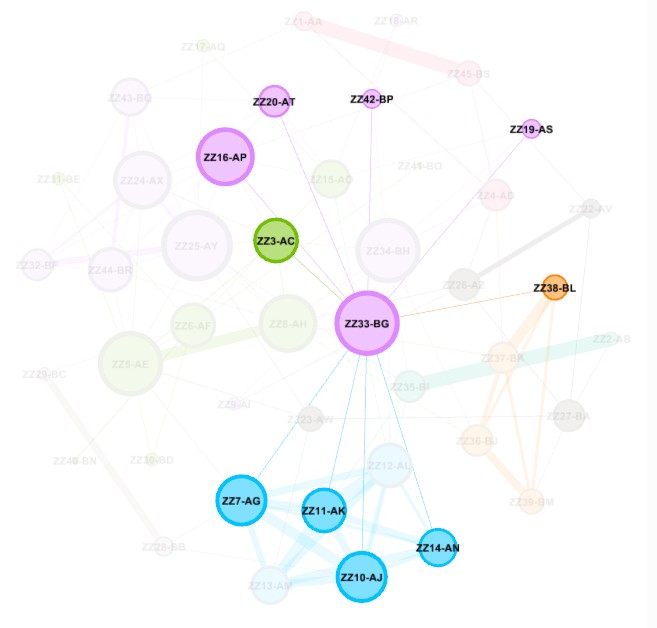
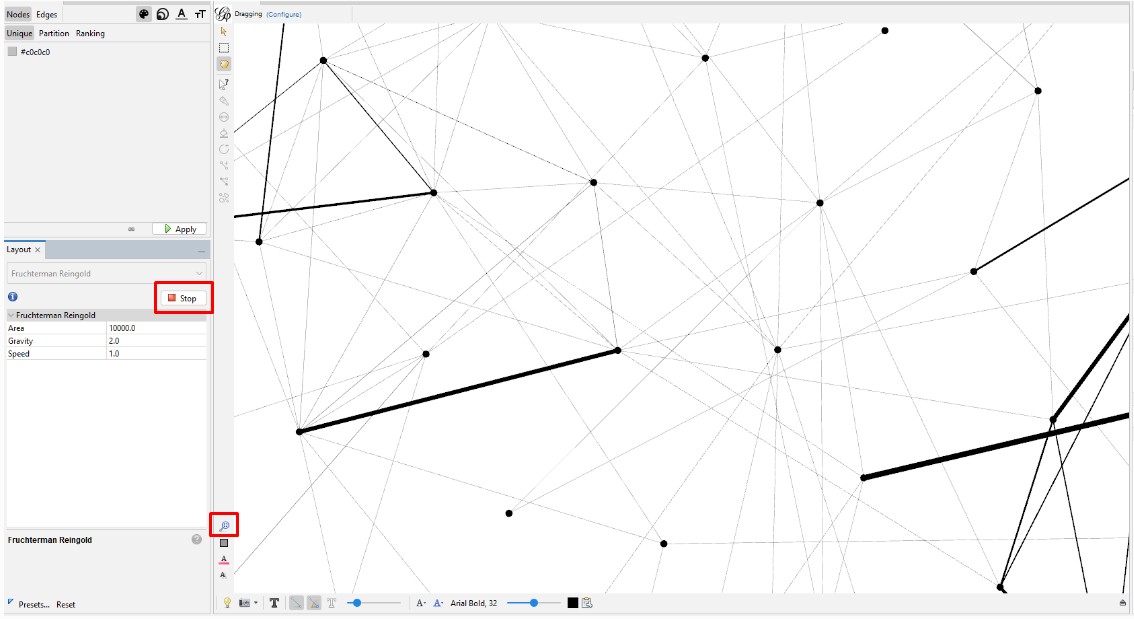
Create and Color Clusters: Now, we need to identify the similar clusters people and give the clusters different colors so that we can start to make some sense of what we see in order to try to gain insight from the graph. In the "Statistics" tab, click on "Run" on the "Modularity" line of the "Community Detection" section. Use the defaults, and click "OK" in the popup window. This is a non-deterministic operation so that if you click "Run" again, it will give a slightly different number. It uses the Louvain community detection algorithm which Dr. David Stumpf reports in his "Graphs for Genealogists" software does a very good job of separating out the different branches of his own family tree.

Back on the left side tool bar, in the "Appearance" section's "Nodes" tab's "Partition" tab, select the attribute of "Modularity Class". Then click "Apply". Since the default is that the icon of an artist's palette was selected, you will see that each cluster/class has a unique color and number (starting from class 0). Once you click "Apply", your graph will show these colors applied to it.

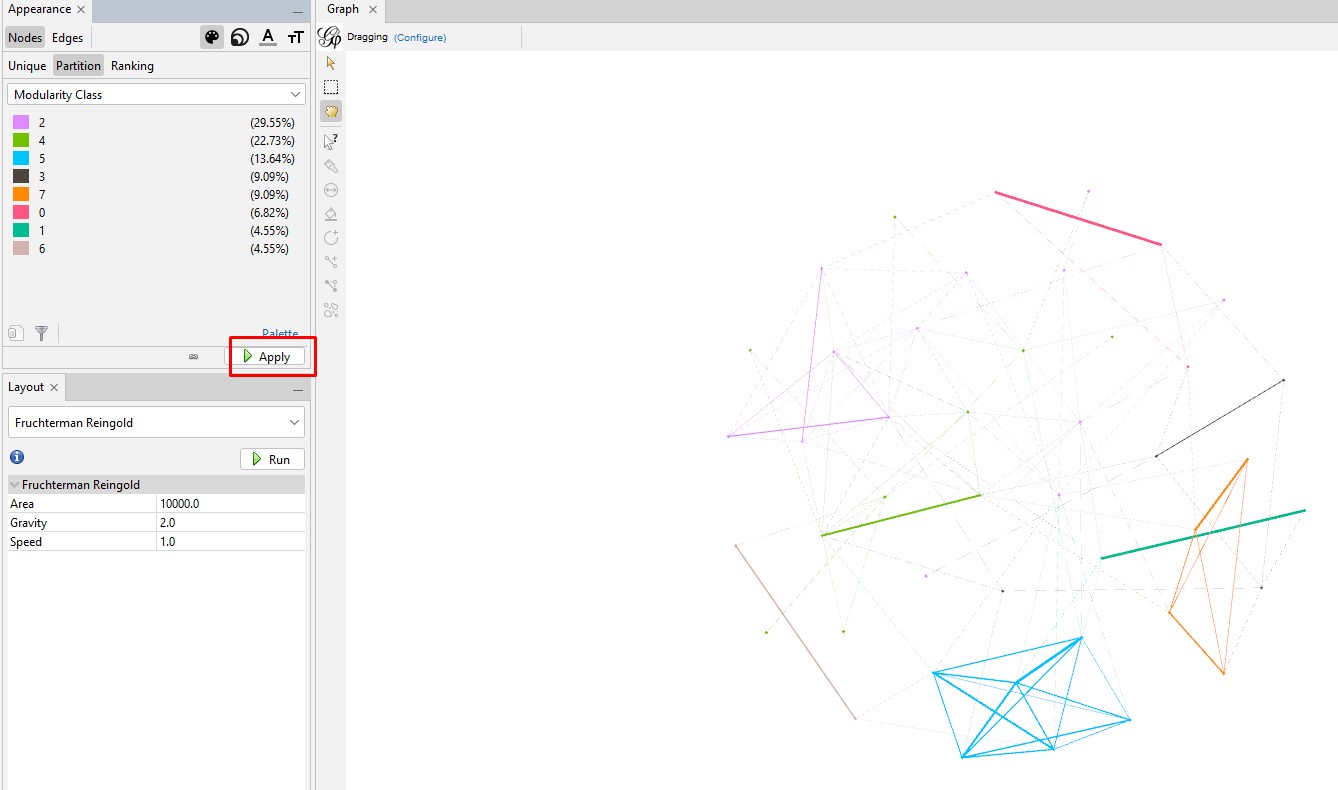
Enhancing the Nodes and Edges: So far, our nodes show and edges show no information other than the connections. We need to know which testers are in which nodes, and it will be useful to set the node's circle size based on how connected that person is to the others in the Autosomal DNA Matrix. In the "Appearance" section, click on the nested circles icon at the top (to the right of the artist's palette icon). Then in the "Ranking" tab, click on the "Degree" attribute in the pulldown list. I change the minimum size to 10 and the maximum to 200. Then click "Apply". Keep in mind that, because these are the most-connected people, even the smallest circles are highly connected.
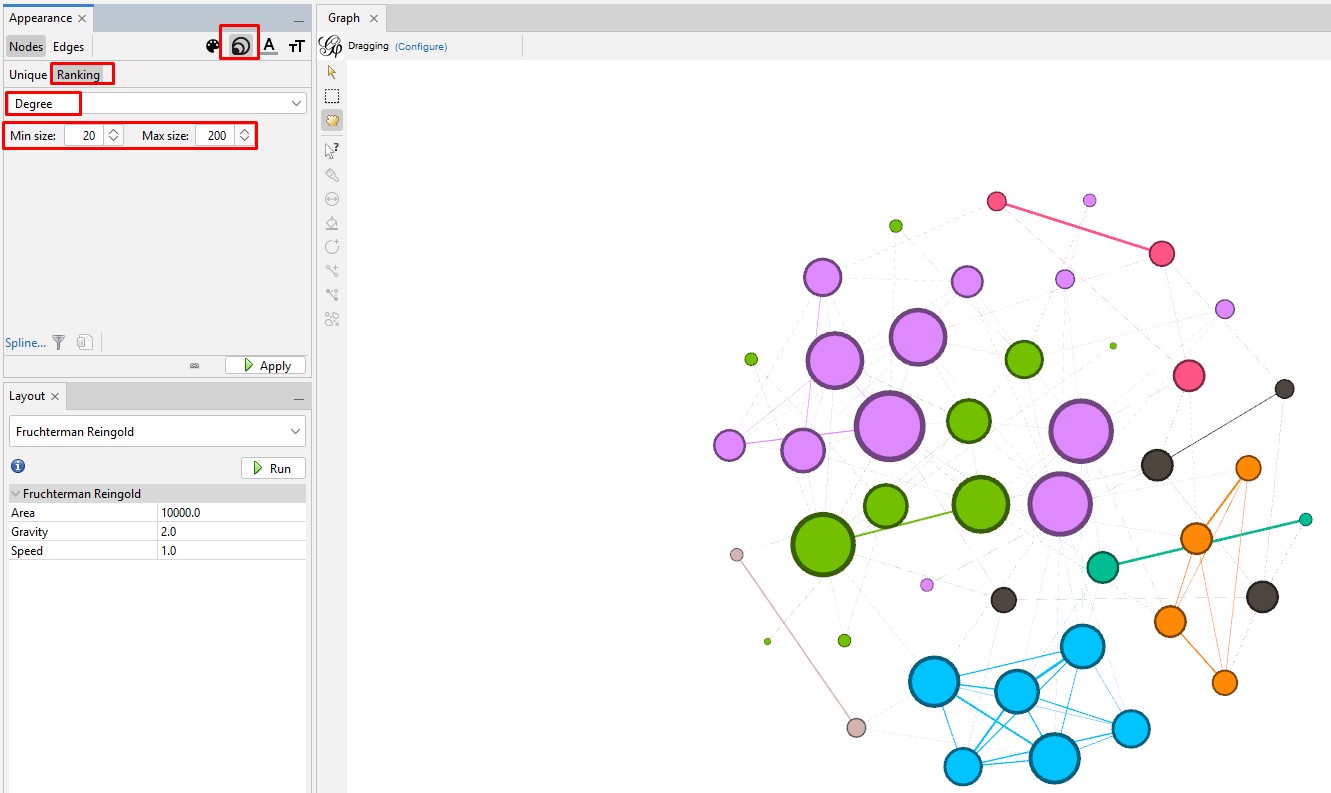
Set the node labels on the tool bar at the bottom of the "Graph" section. At the right end of the bottom toolbar is a stylized up-arrow (looks like a tiny house). Click on that to open the full toolbar. Then click on the "Labels" tab. Click the empty box to check the "Node" section. You can change the font or use the slider to make the labels larger or smaller.
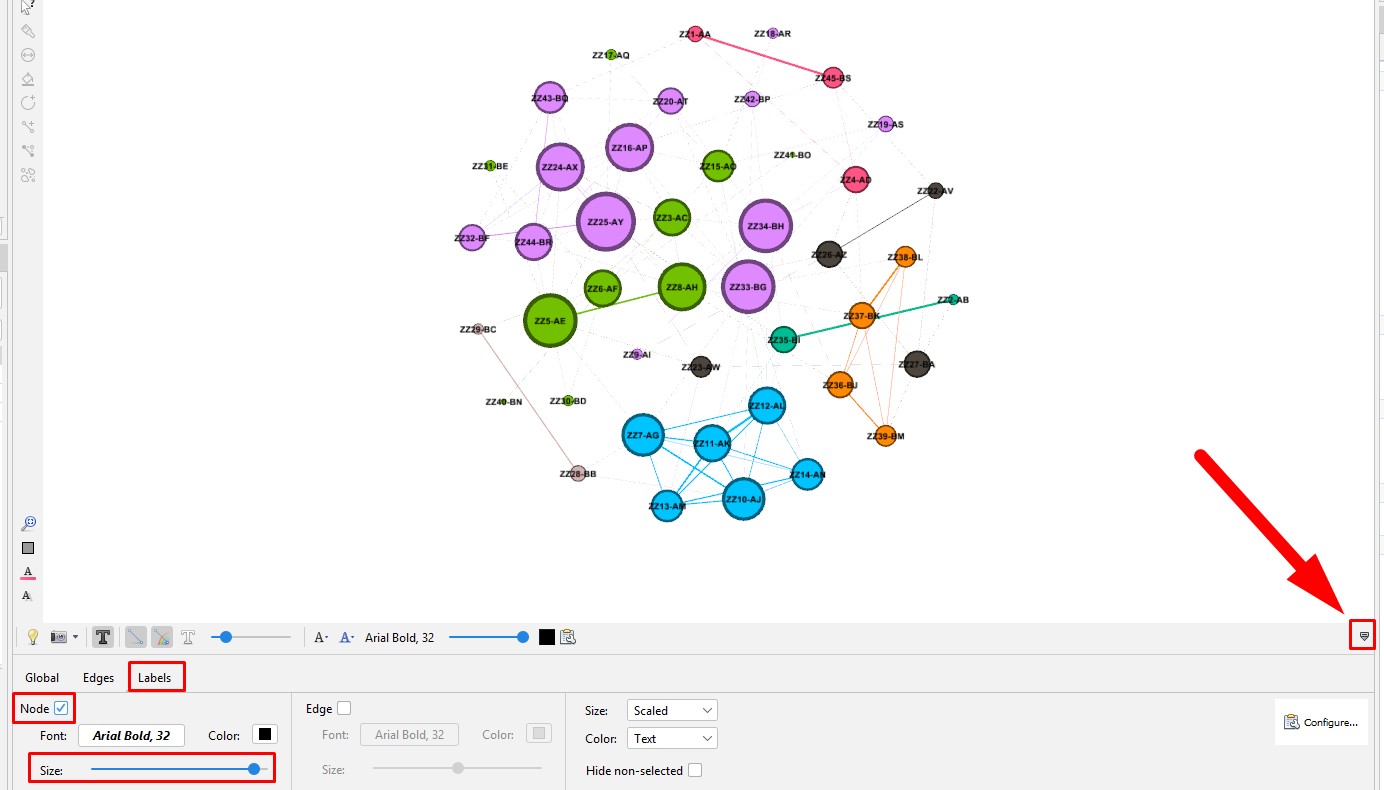
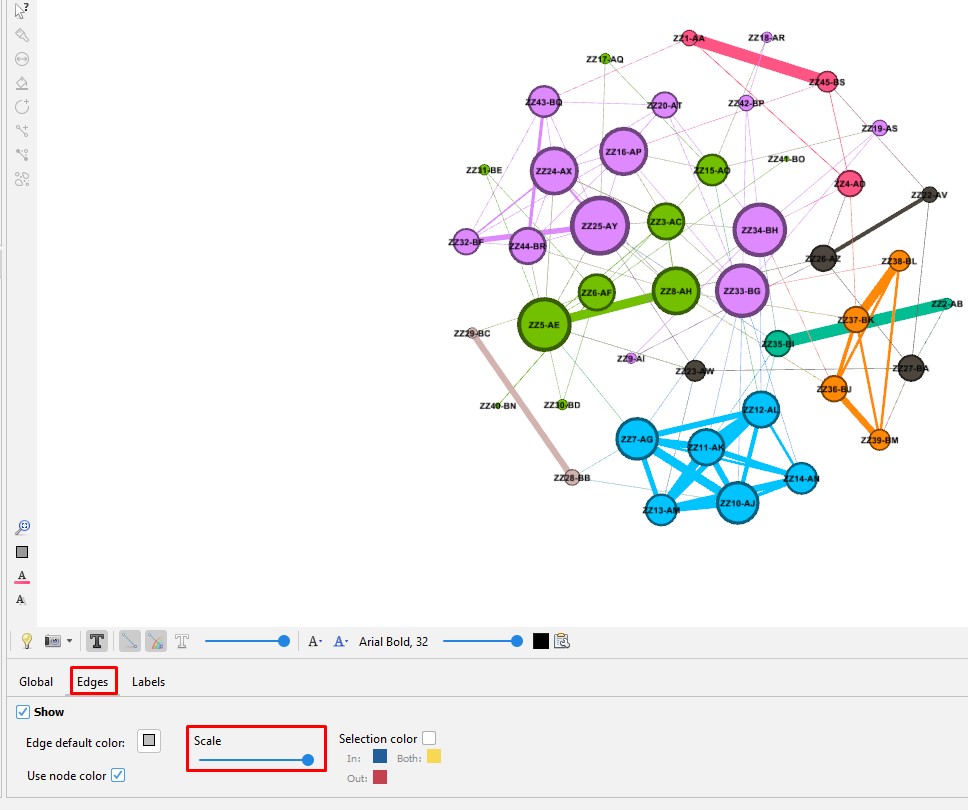
Set the edge size on the tool bar at the bottom of the "Graph" section. Click on the "Edges" tab. Use the slider to make the labels larger or smaller.
|